Summary: The foreach package provides parallel operations for many packages (including randomForest). Packages like gbm and caret have parallelization built into their functions. Other tools like bigmemory and ff solve handling large datasets with memory management.
[table nl=”|”]
Package, Purpose, Benefits
foreach, Workhorse of parallel processing in R. Uses %dopar% to parallelize tasks and returns it as a list of vector of results., Simple syntax||Used in many R packages
gbm, Gradient Boosting Machines build an ensemble of decision trees (one on top of the next) and does a parallel cross-validation, Simple to turn on parallel processing (n.cores).
randomForest, Build ensembles of decision trees. Have to use foreach and the combine function to get true parallelism., Simple to execute and interpret.
caret, Provides a framework of finding optimal parameters by trying lots of models with resampling., Dozens of models work with this package.||Parallel processing baked into the train() function.
bigmemory, Small and fast ecosystem of packages that map large files with a C++ backend.,Doesn’t rely on disk access.
ff, Similar to bigmemory but works with more packages natively. Has a C++ backend but iteratively loads data into memory rather than storing only pointers., Works with more packages natively.||Allows for characters / factor variables.
[/table]
I love R. It is my favorite analytical tool when it comes to exploring data and quickly building models. However, any R user who has worked with real world datasets, memory is always an issue.
In order to get around that, I decided to invest in a powerful PC that will let me fit just about any dataset into memory. However, not everyone can afford that nor can they necessarily afford spinning up a more powerful virtual machine on Amazon Web Services.
Fortunately, modern CPUs have the ability to do parallel processing!
We’ll explore a few packages that handle parallel processing and data that doesn’t fit into memory.
foreach Package
What it does: Provides a new looping construct that allows you to take advantage of multiple cores on a single machine. The main parallel operator is %dopar%. The parallel processing is done on a certain number of cores.
Examples:
library(foreach)
library(doMC)
library(rpart)
registerDoMC(2)
#Make N random selections of the data
random_samples <- foreach(n = 1:1000) %dopar% {
train_idx <-sample(1:150,150, replace=TRUE)
list(train=iris[train_idx,],
test=iris[-train_idx,])
}
model_list<-foreach(i = 1:1000) %dopar% {
mod<-rpart(Species~.,data=random_samples[[i]]$train)
cbind(predict(mod,random_samples[[i]]$test,type="vector"),
random_samples[[i]]$test$Species)
}
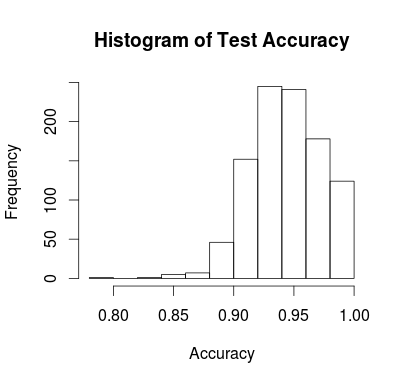
hist(sapply(model_list,
function(x) mean(x[,1]==x[,2])),
main="Histogram of Test Accuracy",
xlab="Accuracy")
How it Works: Developed by the guys at RevolutionAnalytics, the foreach package relies on the doMC package to identify the number of cores available. After setting up your system, you can pretty much call any function inside of %dopar% and the last line variable assignment will be returned.
By default, the results will be returned as a list. You can call .combine = cbind to return the results as a matrix. For example, we could have changed the code above to be like…
model_list<-foreach(i = 1:1000,.combine=cbind) %dopar% {
mod<-rpart(Species~.,data=random_samples[[i]]$train)
pred <- predict(mod,random_samples[[i]]$test,type="vector")
matched <- pred==as.numeric(random_samples[[i]]$test$Species)
}
hist(colMeans(model_list),
main="Histogram of Test Accuracy",
xlab="Accuracy")
This code now returns a matrix which you can use colMeans on to generate the accuracy metric. However, because the test set size can vary, this probably isn’t the best approach. This just highlights how important it is to think through your process before making it parallel!
gbm: Gradient Boosted Machines
What it does: Gradient Boosted Machines creates an ensemble of decision trees that build on top of each other. Each tree predicts the error of the next tree. When combined this ensemble performs extremely well.
Example:
library(gbm) gbm_model <- gbm(Species~., data=iris, n.trees=5000, cv.folds=10,n.cores=8) gbm_model # gbm(formula = Species ~ ., data = iris, n.trees = 5000, cv.folds = 10, # n.cores = 8) # A gradient boosted model with multinomial loss function. # 5000 iterations were performed. # The best cross-validation iteration was 3032. # There were 4 predictors of which 4 had non-zero influence.
How it Works: The gbm package relies on the built-in parallel package. The search for the best decision tree is done across the n.cores. GBM uses cross-validation to identify the best number of decision trees (either using the training or testing error).
randomForest (+ foreach)
What it Does: The randomForest package creates an ensemble of decision trees and their average vote is your prediction. By default, the randomForest package is single threaded (i.e. operates on one processor in one consecutive execution). However, the brilliance of the foreach package allows you to create small forests and then combine them by using the combine function from the randomForest package.
Example:
# Random Forests ####
library(randomForest)
library(doMC)
registerDoMC()
rf <- foreach(ntree=rep(100, 6), .combine=combine, .multicombine=TRUE,
.packages='randomForest') %dopar% {
randomForest(x=data,
y=data_classes,
ntree=ntree)
}
How it Works:
- Using the foreach package, we set up six instances of 100 trees.
- We then call the
.combineparameter and use thecombinefunction from therandomForestpackage. - The combine function just adds the forest from one randomForest object to another, which is perfect for building in parallel.
- The remainder of the code is the creation of a randomForest model.
While it’s not as seamless as GBM’s n.cores parameter, it’s a great system to quickly build large forests.
caret Package
What it does: The caret package provides a “train” function that creates a standardized framework for parameter tuning. Using
Example:
library(caret)
library(doMC)
registerDoMC(cores = 2)
tctrl <-trainControl(method="cv",number=10,repeats=1000)
randforest_model <- train(Species~., data=iris, method="rf",
trainControl =tctrl)
randforest_model$results
# mtry Accuracy Kappa AccuracySD KappaSD
# 1 2 0.9500845 0.9240721 0.02430342 0.03766632
# 2 3 0.9511139 0.9255973 0.02390644 0.03704774
# 3 4 0.9496979 0.9234511 0.02243980 0.03485773
We register two cores using registerDoMC. The random forest (method=”rf”) model uses 10-fold cross-validation and repeats the 10 folds 1,000 times.
The results of the model show that when you randomly select two variables (mtry==2) and use them in the model, you end up with the highest accuracy (0.9511).
How it Works: caret relies on the doMC package and registerDoMC to identify how many cores are available. As the caret package explores the different parameters, it will send the different models to different cores. Based on some experimentation, it looks like it also sends the cross-validation (like in gbm) to the different cores.
Other Packages
There are a few other packages that have a small ecosystem of supported features that are worth exploring or at least knowing about.
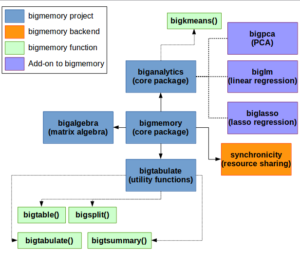
bigmemory Package
What it does: Designed to work with large numeric matrices that may fit in memory but doesn’t leave room for actually working with the data. The bigmemory package only works with numeric variables and converts any factor into its numeric underlying value.
Examples: I haven’t actually been able to get an example to work.
How it Works: Developed by Michael Kane and others, bigmemory relies on a C++ backend that maps sections of the data to a set of pointers. The usual R commands on matrices and data.frames (like df[r,c]) are overridden and translated to C++ functions (like GetMatrixElements).
ff Package
What it does: Similar to bigmemory, but relies on loading chunks of the data into memory and allows many (but not all) packages to take advantage (for example randomForest fails).
Examples:
library(ff) ffdata <- read.csv.ffdf(file="~/Downloads/2008.csv",header=T) lm(LateAircraftDelay~Year+Month+ArrTime+Distance,data=ffdata) rpart(LateAircraftDelay~Year+Month+ArrTime+Distance,data=ffdata) #randomForests doesn't work natively with ff randomForest(LateAircraftDelay~Year+Month,data=ffdata)
Bottom LineThe foreach and doMC packages are the core of R’s parallel processing and a few packages like GBM, randomForest, and caret have built-in parallel processing capabilities.